Understanding how eukaryotic chromosomes are maintained but sometimes changed
Chromosomal DNA contains the blueprint of life. Thus, chromosomes need to be maintained even though they undergo complex reactions such as replication and segregation as cells proliferate. However, gross chromosome rearrangements (GCRs) that alter the size or number of chromosomes may occur in rare cases. GCR can cause genetic diseases such as cancer and cell death. Interestingly, chromosome maintenance and GCRs arise by the action of enzymes produced in the same organism. Our research group aims to elucidate the molecular mechanisms of chromosome maintenance and GCRs and unravel the mystery of life, the “struggle” between the two.
FEATURE
We aim to discover new biological phenomena and elucidate their molecular mechanisms.
We aim to develop new research areas through original research.
Our research contributes to developing new diagnoses and medical care for cancer, autism, etc.
RESULTS
Research progress
Understanding the mechanism of gross chromosomal rearrangements (GCRs) mediated by repetitive DNA sequences at centromeres
(1) The formation of isochromosomes that are mediated by centromere repeats
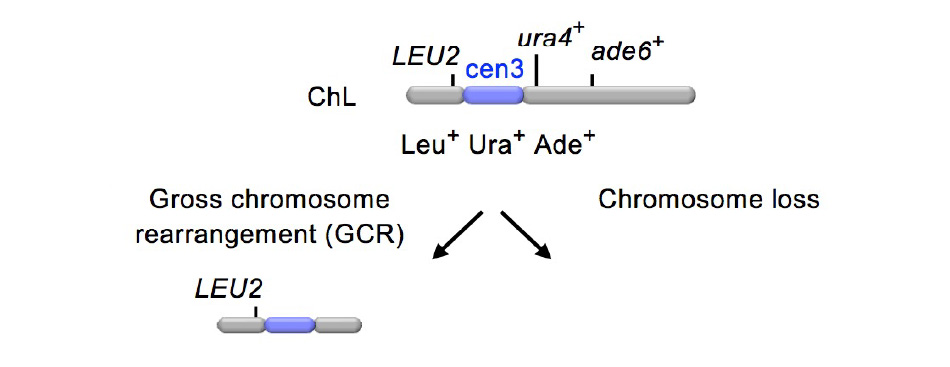
Using the fission yeast Schizosaccharomyces pombe, our research group has developed an experimental system that can determine the spontaneous rate of gross chromosomal rearrangements (GCRs) and the structure of the GCR products. The centromere is a chromosomal region essential for faithful segregation of chromosomes and consists of repetitive DNA sequences. We succeeded in detecting the isochromosomes that are formed by GCR using centromere inverted repeats.
(2) Suppression of GCR by Rad51-dependent homologous recombination
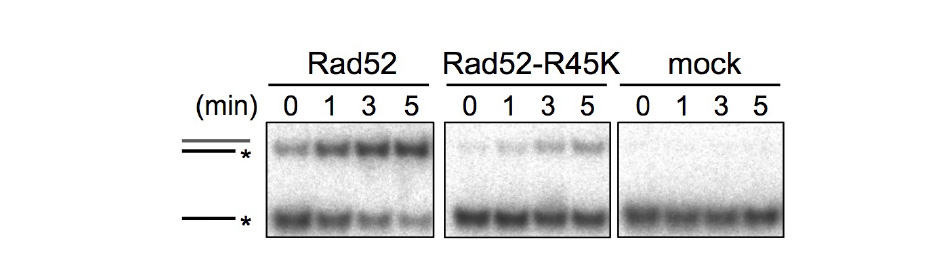
Rad51 is an evolutionally conserved protein that facilitates the pairing of homologous sequences. Our group has shown that Rad51-dependent homologous recombination promotes conservative recombination between centromere repeats, thereby suppressing isochromosome formation. Interestingly, Rad51-dependent homologous recombination occurs more actively in the centromere region than in other chromosomal regions. Rad52, which catalyzes Rad51-independent homologous pairing such as single-strand annealing (SSA), causes isochromosome formation.
(3) Heterochromatin suppresses GCR by transcriptional repression
In nuclei, DNA is wrapped around histone octamers forming nucleosomes. The nucleosome is the basic unit of chromatin structure which can be classified into two forms: highly condensed heterochromatin and relaxed euchromatin structures. Chromosomal regions containing repetitive sequences, such as centromeres and telomeres, are transcriptionally repressed by forming heterochromatin structures. Our group has shown that disruption of heterochromatin structure activates transcription at the centromere, resulting in isochromosome formation.
Further development
Molecular mechanisms of chromosome maintenance and chromosomal rearrangements
Molecular biology has made great strides in our understanding of individual biological phenomena such as chromosome replication and chromosome segregation. However, how these phenomena interact and harmonize with each other in the cell remains to be determined. The same seems to be true for DNA replication, transcription, recombination, and epigenetic regulation, which are involved in chromosome maintenance and chromosomal rearrangements. Future work will seek to unravel the linkages between these biological reactions to gain a core understanding of chromosomes. We are studying the relationship between conservative recombination and chromosome aberrations and the mechanism by which transcription induces gross chromosomal rearrangements.